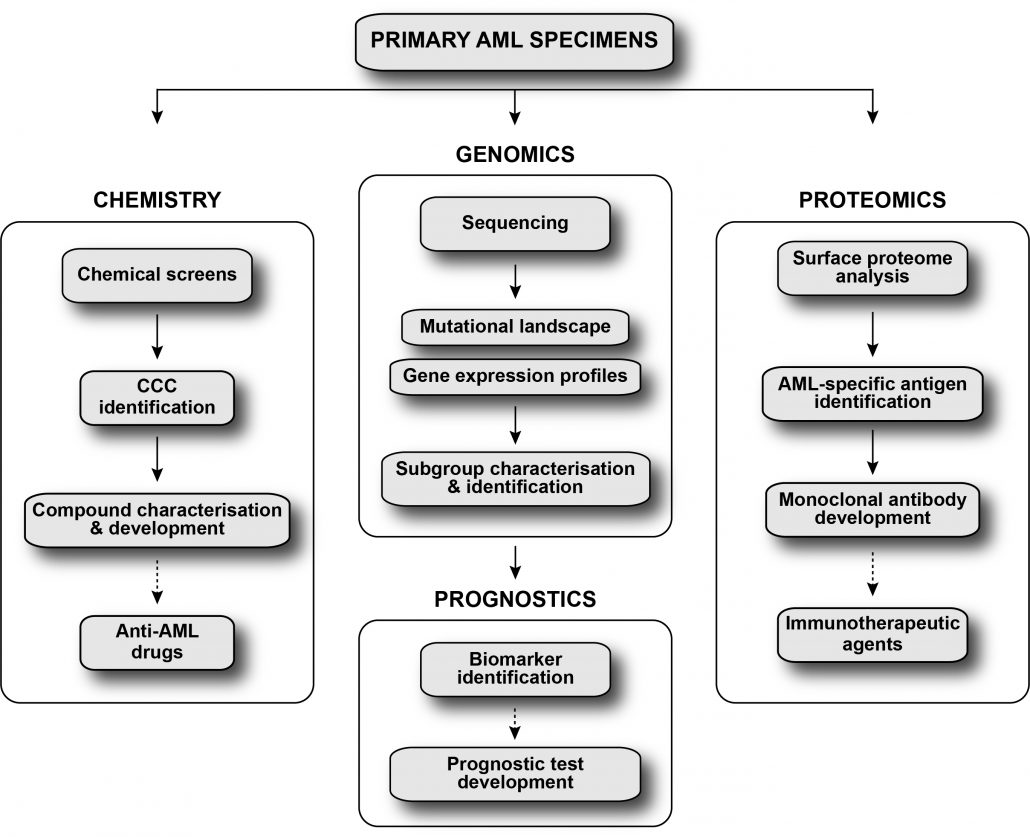
The Leucegene project aims to improve acute myeloid leukemia stratification and to develop novel therapeutic strategies for this disease using an integrated approach.
Acute myeloid leukemia (AML) is diagnosed every year in more than 50,000 individuals in Canada, US and Europe, and is associated with a 5-year overall survival nearing 27%. This highly lethal disease is characterized by several chromosomal aberrations and molecular alterations, which form the basis of the current genetic risk model stratifying AML patients into three categories: favorable, intermediate and adverse. The risk category guides the choice of consolidation therapies, which is straightforward for favourable (chemotherapy) and adverse (stem cell transplantation) risk patients. However, management of patients falling into the intermediate risk category is challenging, as prediction of relapse remains inaccurate. This leads to over- and under-treatment, resulting in serious medical, social and economical consequences. Hence, there is an urgent need for precise prognostic markers that better stratify intermediate-risk patients as well as for novel therapeutic strategies for the treatment of adverse risk AML patients. The approach of the Leucegene project to improve this situation is to build upon our current know-how and add further expertise in functional genomics, medicinal chemistry, proteomics and prognostic test development to identify new prognostic markers and develop novel therapeutic strategies that will improve the management of AML patients.
Identification of novel anti-AML drugs
in collaboration with Anne Marinier’s group
AML is a highly heterogenous disease and thus can be viewed as a multitude of diseases. This partly explains the variable response to first-line therapeutic regimen (cytarabine + anthracycline) observed for AML patients. This project aims to identify new treatment avenues and explore AML subtype-specific sensitivities to drugs by exposing primary human AML specimens to collections of compounds under optimized ex vivo culture conditions supporting leukemia stem cell activity. Response patterns to these compounds are hierarchically grouped to form “Compound Correlation Clusters” or CCCs, representing groups of compounds with similar inhibitory profiles across specimens (Baccelli et al., Blood cancer J, 2017). In close relation to genomic classification of AML subgroups, this approach highlights compounds with interesting therapeutic potential (e.g. targeting specific AML subgroups or AML specimens with high LSC frequency or associated with bad prognosis: Krosl et al., Blood advances, 2021; Moison et al., Blood advances, 2019; Baccelli et al., Cancer cell, 2019; Bisaillon et al., Leukemia, 2019; Simon et al., Clinical cancer research, 2017). The anti-AML activity of these compounds is then validated using both pharmacological and genetic approaches, and in vivo activity assessed through preclinical studies using mouse models. With this strategy, we hope to identify and target the Achilles’ heels of the different AML subtypes.
Genomic characterization of AML subgroups
in collaboration with Josée Hébert’s, Vincent-Philippe Lavallée’s and Sébastien Lemieux’s groups
Refining AML patients stratification is a major challenge and represents one of the keystones of both diagnosis and therapy improvement. As part of the Leucegene project, our group sequenced close to 700 primary AML specimens (DNA and/or RNA) selected to best represent the genetic diversity of the disease. Using this unique dataset, we are developing multidisciplinary projects combining -omics, molecular biology and innovative analysis methods, to dissect the determinants that shape AML variability (e.g. mutations, structural rearrangements, differentially expressed genes). More specifically, our approach involves the comparison of data from specimens of a given genetic subgroup with sequencing results from the other specimens of the cohort, with a focus on gene/mutation signatures comprising a limited number of candidates, rather than on more conventional global clustering. Using this strategy, we confirmed all known subgroup-specific mutations in AML, and more importantly, we identified new ones allowing patient reclassification (Simon et al., Blood, 2020; Lavallée et al.: Leukemia (2018), Blood (2016 (2)), Leukemia (2016), Nature Genetics (2015), Blood (2015); Lehnertz et al., Blood, 2017). Leveraging our unique dataset, we aim to better characterize rare AML subgroups and identify new ones, to ultimately propose new diagnostic and prognostic markers.
Development of immunotherapeutic agents for AML
in collaboration with Philippe Roux’s group
Antibody-based immunotherapies are attractive therapeutic options as they produce antitumor activity with reduced systemic toxicity compared to conventional therapies. While this approach holds great promise, its development inexorably depends on the identification of reliable cell surface antigens selectively expressed by cancer cells. Historically, the identification of tumor cell surface antigens has been challenging due to the characteristic hydrophobicity of plasma membrane proteins, which impedes their purification by biophysical methods. Furthermore, compared to solid tumors, antibody-based immunotherapy remains underexploited for AML, likely due to the limited availability of AML-specific targetable antigens. Our group has built an integrated pipeline combining transcriptome and surface proteome analysis of primary human AML specimens to identify novel antigens for this disease. This approach already identified many antigens for the different AML subgroups. Therapeutic antibodies targeting these antigens are currently being developed and tested for their ability to eliminate primary human AML cells in culture and eventually in vivo.
AML prognostic tests development
in collaboration with Guillaume Richard-Carpentier and Josée Hébert’s group
The Leucegene team is currently working to develop novel prognostic tests to better orient treatment of AML patients. Using RNA sequencing data from the Leucegene cohort and clinical data from the Banque de cellules leucémiques du Québec, a first prognostic marker, HMGA2, was identified (Marquis et al., Blood cancer Journal, 2018). Josée Hébert’s pre-clinical laboratory at Hôpital Maisonneuve-Rosemont developed and validated a pronostic test for this marker that predicts poor clinical outcome in AML. With this pipeline and expertise in place, the Leucegene team is currently identifying additional biomarkers with prognostic value to better stratify intermediate risk patients and improve treatment outcome.